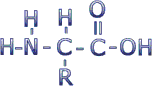A Musical Introduction to Protein Structure
Proteins are composed of subunits called amino acids. Amino acids have the general chemical structure below (C = carbon, H = hydrogen, O = oxygen, N = nitrogen). These atoms are clustered into functional groups. Two of these functional groups define the amino acids: the amino group (the NH2 on the left) and the carboxyl group (the COOH on the right).
There are 20 different amino acids incorporated into proteins. All amino acids have the same general structure, but each has a different R-group -- the chemical group represented by the designation "R" in the diagram above. The carbon atom to which the R group is connected is called the alpha carbon. Click here to see a table of amino acid R-groups from the Molecular Biology Web Book.
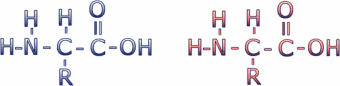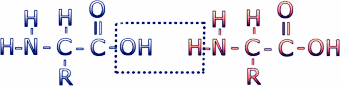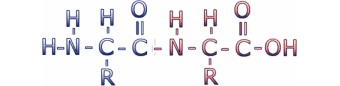
Amino acids are connected to make proteins by a chemical reaction in which a molecule of water is removed, leaving two amino acids residues (i.e. what's left when the water is removed) connected by a peptide bond. Connecting multiple amino acids in this way produces a polypeptide. Click on the image to see an animation of this process.
This reaction leaves the C of the carboxyl group directly linked to the N of the amino group. This linked group of atoms (CONH) is called the peptide bond. Polypeptides can be thought of as a string of alpha carbons alternating with peptide bonds. Since each alpha carbon is attached to an R-group, a given polypeptide is distinguished by the sequence of its R-groups. In the protein data bases, each R-group is represented by a single letter of the English alphabet.
A = alanine
C = cysteine
D = aspartate
E = glutamate
F = phenylalanine
G = glycine
H = histidineI = isoleucine
K = lysine
L = leucine
M = methionine
N = asparagine
P = prolineQ = glutamine
R = arginine
S = serine
T = threonine
V = valine
W = tryptophan
Y = tyrosineAmino acid R-groups can be divided into four families: water-insoluble (hydrophobic) , water-soluble (hydrophilic) , positively charged and negatively charged (both very hydrophilic). The names of the amino acids in the table above have been color coded according to family resemblances. The small amino acid glycine is a special case as it effectively has no R-group.
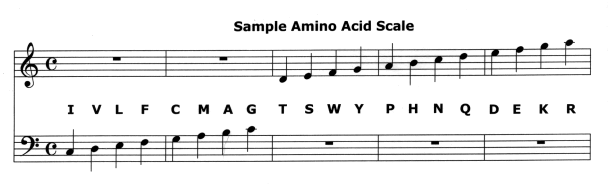
The twenty amino acids can be assigned to a musical scale, for example the C-major scale below. A number of different criteria can be used in selecting pitches to represent the amino acids. In this scale, the amino acids are ordered by their relative hydrophobocity. Some of the choices below are arbitrary, for example Q, N, D and E all have the same hydrophobicity values.
Click here to play this Amino Acid Scale.
The duration of each note varies with the number of DNA codons associated with the amino acid. The DNA codons are represented by a harp playing the three bases of each codon under its amino acid. The last three codons to sound are stop codons and do not correspond to any amino acid.
The linear order of amino acids in a polypeptide is called its primary structure. Primary structure is represented in the protein data bases by a string of the single letters, like a long word or sentence. The order of letters is the order in which the amino acids were strung together when the polypeptide was synthesized. This order is specified by genetic information in the form of a string of DNA codons: sets of three bases from the four base DNA alphabet. Click here to see a Table of the Genetic Code. The letters below represent the sequence of human calmodulin. Calmodulin is a calcium-binding protein; the four calcium binding sites are underlined. Click here to see the coding DNA for Calmodulin.
ADQLTEEQIAEFKEAFSLF
DKDGDGTITTKELGTVMR
SLGQNPTEAELQDMINEV
DADGNGTIDFPEFLTMMARK
MKDTDSEEEIREAFRVF
DKDGNGYISAAELRHVMTN
LGEKLTDEEVDEMIREA
DIDGDGQVNYEEFVQMMTAKAssigning pitches to amino acids allows us to play the tune generated by the sequence of amino acids in a polypeptide. Because humans communicate using speech, our brains are very good at recognizing sound patterns. Click on the link below to hear the patterns represented in the primary sequence of calmodulin. played using a major scale like that illustrated above.
Calmodulin tune + Calcium binding sites
The entire sequence is played through by the same synthesized voices used in the scale above, with vibraphone entering four times to play the Calcium binding sites. When an amino acid is repeated, the note is sustained until the next amino acid appears in the sequence. The protein sequence is accompanied by harp playing the DNA codons. After you have listened to the tune a couple of times, see if you can follow the written sequence as the tune plays out.
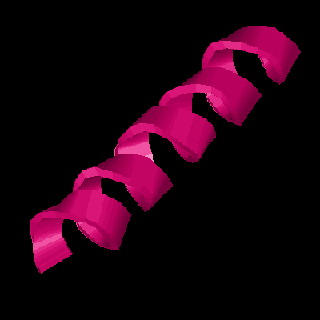
Proteins fold up into complex three dimensional shapes that give them the ability to interact with other molecules -- Calmodulin is shaped to recognize and bind Calcium. The first level of protein folding is called secondary structure and consists of very regular folding patterns stabilized by weak interactions between the atoms of the peptide bond. Three common types of secondary structure are alpha helix, beta strands, and turns.
Alpha helix looks like a spring (and is springy), beta strands are folded back and forth like an accordion pleat and may align with other beta strands to form a beta sheet, and turns are just a simple bend in the protein chain. The sequence below is marked to show what parts of the amino acid sequence of calmodulin fold in these different ways. In the musical example that follows the sequence, these different folds are represented by the different instruments listed. The "DNA harp" plays throughout, but the flute begins with the first alpha helix below. The vibraphone continues to play the Calcium binding sites.
ADQL(TEEQIAEFKEAFSLF)
D<KD>GDG[TI]T(TKELGTVMR)
<SLG>QNPT(EAELQDMINEV)
DADGNGTI[DF](PEFLTMMARK)
(MKDTDSEEEIREAFRVF)
D<KD>GNGY[I]S(AAELRHVMTN)
<LG>EKLT(DEEVDEMIREA)
DIDGDGQ[V]N(YEEFVQMMT)AKCalmodulin Secondary Structure
() = alpha helix (flute)
[ ] = beta strand (tubular chime)
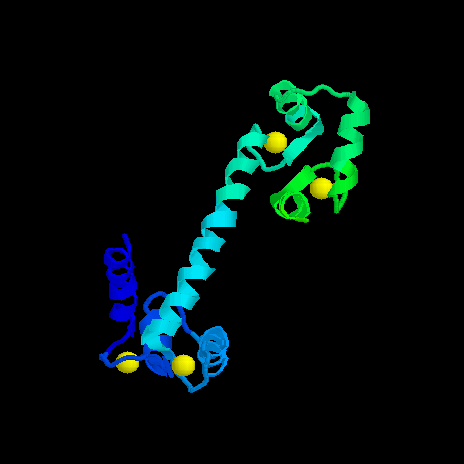
<> = turn (crystal bells)Note that each Calcium binding site is bracketed by two regions of alpha helix. These regions of secondary structure compose part of a larger, more complex folding pattern call the protein tertiary structure. The image below represents the tertiary structure of Calmodulin. Locate the helical ribbons, the flat beta strands and the turns in this figure. The four yellow balls represent the Calcium bound at the two ends of this dumbbell shaped molecule. In this image, the sequence begins with the lime green section at the upper right and ends with the blue helix at the lower left.
Tertiary structure is maintained by interactions of R-groups. The relative placement of the different R-groups determines whether a given section of a protein will form alpha-helix, a beta-strand, or some other folding configuration. Hydrophobic (water insoluble) groups tend to hang out together, hydrophilic (water soluble) groups tend to hang out together, and the positive and negative R-groups may attract each other. Generally hydrophobic R-groups will line up along one edge of a helix or in the interior of a globular region.
The next musical example illustrates how the hydrophobic and hydrophilic R-groups are distributed in the protein. The lower and higher solubility R groups are represented by different voices so that the musical phrases are divided into two groups: one group consists of the hydrophobic amino acids (lower pitches) and the other group consists of the hydrophilic (higher pitches) amino acids. Among the hydrophilic amino acids, the charged ones have been asigned the highest pitches. You can think of the folding structure of a protein as a hydrophobic core decorated or overlaid with hydrophilic surfaces. The lower tones in these duets define the structural core of the protein.
Two "choruses" sing the full sequence, while two vibraphones play the Calcium binding sites.
In the next musical example, you can hear how the hydrophobic and hydrophilic amino acids are arrayed in the various regions of secondary structure. In this version, the soluble amino acids of the helices that dominate this protein have been accentuated by the flute.
Obviously the major scale used in these examples is not the only possible way to represent the 20 amino acids that compose proteins, nor is it necessary to stick to the tempered western scale. Although it seems natural to me to use the lower pitches for the protein core, the scale could be inverted or even reorganized altogether using different features of the amino acids to determine pitches and note durations. As an example, listen to the piece below, also based on calmodulin and using the same voices as the sample immediately above. However, in this case, all amino acids with similar solubilities were assigned the same pitch. Does this treatment allow you to hear patterns in this protein that you did not hear before?Evolutionary Improvisation
Musical transformations are also useful for demonstrating evolutionary improvisation -- the minor changes that occur in the proteins of two lineages after they become genetically discontinuous, i.e. when new species form. The next example demonstrates some of the changes that have occurred within the mammalian beta-globins, which form part of the structure of hemoglobin. Four mammals have been selected that represent the major lines of mammalian descent. The tupaia or tree shrew is thought to be similar to the primitive mammals at the root of the mammalian tree. African elephants represent the Afrotheria -- a diverse lineage of mammals that came out of Africa. Sumatran tigers represent the Laurasiatheria -- a group that arose on the ancient northern supercontinent that included Europe, Asia, and North America. Humans represent the unnamed group that includes the primates and the rodents.
In this piece, the 146 amino acids of beta globin have again been separated into hydrophobic and hydrophic groups, with harp, bass and guitar representing the lower octave of hydrophobic R-groups (I V L F C M A G), and flute representing the the upper notes of the hydrophilic groups. The sequence plays through three times, with different species contributing their improvisational changes.Think of an ensemble with four musicians: tree shrew, human, Sumatran tiger and African elephant. The piece begins with the tree shrew playing both the upper and lower voices with flute and harp respectively. At the point indicated on the Beta Globin Sequences chart, tree shrew hands off to human, who plays bass in a duet with tree shrew's harp, and who takes over the flute line.
On the second iteration of the tune, tiger enters, plays guitar in trio with tree shrew and human and also takes over the upper flute part. Again about halfway through, elephant enters, adding harp to make a "hydrophobic quartet," and in turn takes the flute line to the end of the sequence. In all the duets, trios and quartets, sequence divergence is heard as a chord; otherwise the species play in genetic unison.
On the final iteration, tree shrew and human play both the hydrophobic and hydrophilic lines of the full sequence on their respective instruments. As you listen to the last section, which part of the sequence has diverged more between these two species: the hydrophobic core or the hydrophilic surface notes?Big ProteinsMany proteins, like calmodulin or beta globin, are relatively small -- around 150 - 300 amino acid residues. Some proteins, like hemoglobin, are constructed of a cluster of subunits. Multichain proteins like hemoglobin are said to have quaternary structure.
However, some proteins are truly huge. The largest single protein is probably the muscle protein titin, which contains 27,000 amino acid residues in a single chain. The CFTR (Cystic Fibrosis Transmembrane conductance Regulator) protein that forms a Chloride channel in cell membranes has 1408 amino acids. Another large protein is the Huntington's Disease protein, with 3144 amino acids.
Large proteins like this are divided into a number of structural domains, each of which may include specific recognition sites that allow it to interact with other proteins. It is possible to get a sense of the large scale structure of such proteins by playing through them rapidly. Because of the large file size of the full sequence, you may want to play only the shorter sample sequence.
Huntington's Protein Sample Sequence
(includes PolyQ region and first HEAT repeat domain. Playing time 2:00)Huntington's Protein Full Sequence
(Large file size. Playing time 13:15)Four voices speak in the readthrough of this sequence. The low and high solubility amino acids are read by koto and a sitar/flute combination respectively. The HEAT repeat regions are marked by the entrance of crystal chimes. This protein sequence is set in a pentatonic scale.
Sources:
Sequence information from Swiss Prot
Images from the Protein Data Bank
Secondary structure figures from the
Molecular Biology Group (Cornell University)